Simulation with Docker
Creating and saving the dataset as a file is made simple by utilizing an existing Docker image available on DockerHub. Begin by pulling one of the existing images using the following command:
docker pull adku1173/acoupipe:latest-full
This image encompasses the simulation source code along with an up-to-date version of Acoular, AcouPipe, and Tensorflow.
To initiate the dataset generation, run a Docker container with the following command:
SPLIT='training'
#SIZE=<enter desired size of the dataset>
NTASKS=<enter the number of parallel tasks> # should match the number of CPUs on the host
docker run -e NUMBA_NUM_THREADS=1 -it --user "$(id -u)":"$(id -g)" -v $(pwd):/app adku1173/acoupipe:latest-full python /app/main.py --tasks=$NTASKS --size=$SIZE --split=$SPLIT --features 'csm'
Take note that the current user on the host is specified as the user of the Docker environment using the additional argument --user "$(id -u)":"$(id -g)". Running the container as a root user is not recommended. It’s essential to bind a directory where the dataset files are stored to the container. With the command -v $(pwd):/app, the current working directory on Linux or macOS hosts is bound.
The simulation is capable of running on multiple CPU threads concurrently, enhancing computation speed. Users can specify the exact number of threads using the --tasks` argument. To prevent thread overloading caused by multiple parallel processes, we enforce Numba to use only one thread by passing the environment variable NUMBA_NUM_THREADS=1. For demonstration purposes, the script calculates the training split.
After starting the main script, a progress bar should appear that logs the current simulation status:
1%|█▍ | 83/10000 [01:04<1:40:35, 1.64it/s]
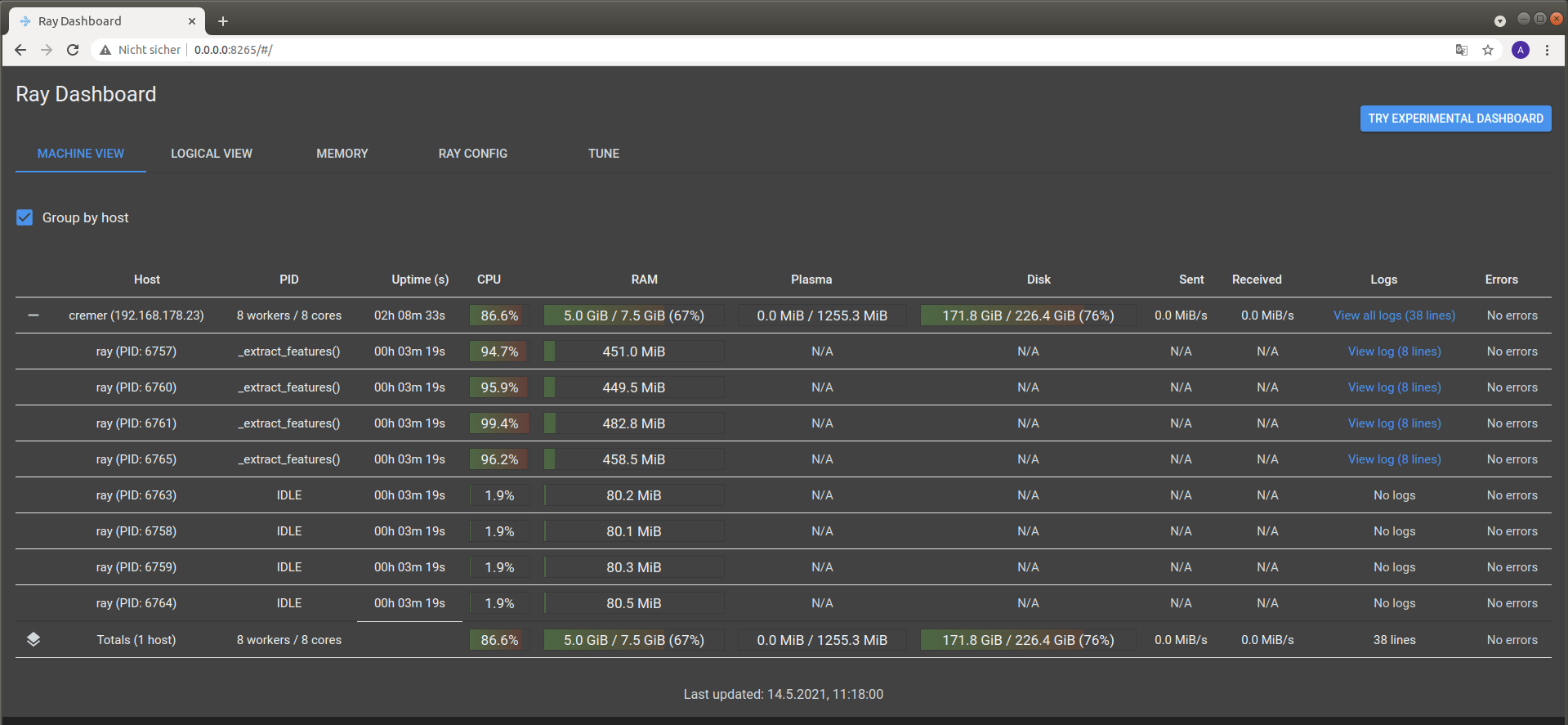
It is possible to view the CPU usage in a dashboard application served by the Ray API. One should find the following output at the beginning of the simulation process when running the simulation on multiple CPU threads
2021-05-14 08:50:16,533 INFO services.py:1267 -- View the Ray dashboard at http://0.0.0.0:8265
It is necessary to forward the corresponding TCP port with docker run -p 8265:8265 ... at the start-up of the container to access the server serving the dashboard.
One can open the dashboard by accessing the web address http://0.0.0.0:8265 which should display the following web interface
The main.py script has some further command line options that can be used to influence the simulation process:
usage: main.py [-h] [--dataset {DatasetSynthetic,DatasetMIRACLE}] [--name NAME]
[--features {time_data,csm,csmtriu,sourcemap,eigmode,spectrogram,loc,source_strength_analytic,source_strength_estimated,noise_strength_analytic,noise_strength_estimated} [{time_data,csm,csmtriu,sourcemap,eigmode,spectrogram,loc,source_strength_analytic,source_strength_estimated,noise_strength_analytic,noise_strength_estimated} ...]]
[--mode {welch,wishart,analytic}] [--format {tfrecord,h5}] [--f F [F ...]] [--num NUM]
[--split {training,validation,test}] --size SIZE [--start_idx START_IDX] [--tasks TASKS]
[--head HEAD] [--log]
options:
-h, --help show this help message and exit
--dataset {DatasetSynthetic,DatasetMIRACLE}
Which dataset to compute. Default is 'DatasetSynthetic'
--name NAME filename of simulated data. If 'None' a filename is given and the file is
stored under './datasets'
--features {time_data,csm,csmtriu,sourcemap,eigmode,spectrogram,loc,source_strength_analytic,source_strength_estimated,noise_strength_analytic,noise_strength_estimated} [{time_data,csm,csmtriu,sourcemap,eigmode,spectrogram,loc,source_strength_analytic,source_strength_estimated,noise_strength_analytic,noise_strength_estimated} ...]
Features included in the dataset. Default is the cross-spectral matrix 'csm'
--mode {welch,wishart,analytic}
Calculation mode of the underlying Cross-spectral matrix. Default is 'welch'
--format {tfrecord,h5}
Desired file format to store the datasets. Defaults to '.h5' format
--f F [F ...] frequency or frequencies included by the features and labels. Default is 'None'
(all frequencies included)
--num NUM bandwidth of the considered frequencies. Default is single frequency line(s)
--split {training,validation,test}
Which dataset split to compute ('training' or 'validation' or 'test')
--size SIZE Total number of samples to simulate
--start_idx START_IDX
Start simulation at a specific sample of the dataset. Default: 0
--tasks TASKS Number of asynchronous tasks. Defaults to '1' (non-distributed)
--head HEAD IP address of the head node in the ray cluster. Only necessary when running in
distributed mode.
--log Whether to log timing statistics. Only for internal use.